NCPI FHIR Implementation Guide - Local Development build (v0.2.0) built by the FHIR (HL7® FHIR® Standard) Build Tools. See the Directory of published versions
Example Research Study Baylor Cmg
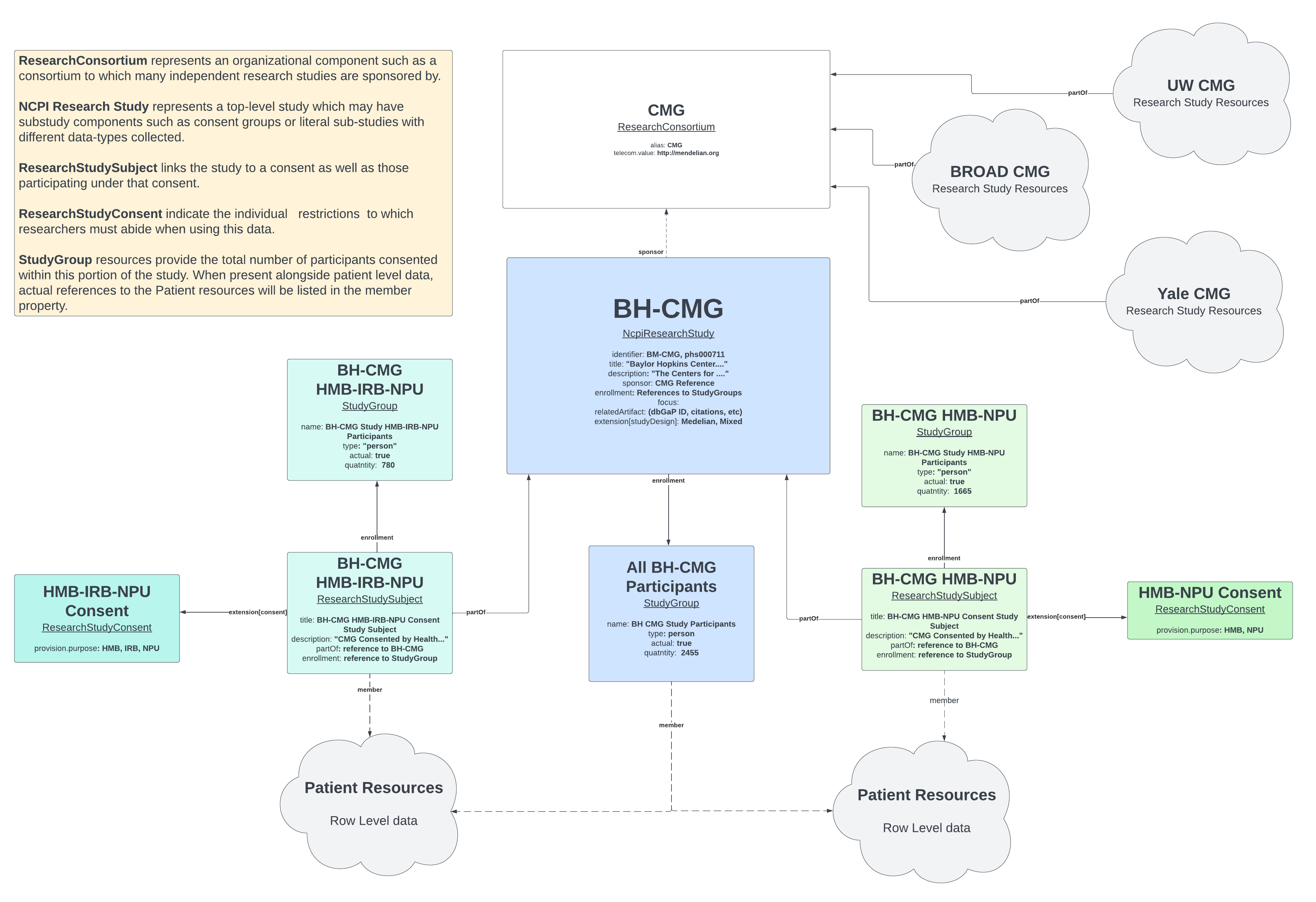
Research Study details can be represented in FHIR using a number of key Profiles defined within the NCPI Implementation Guide. Together, these profiles can be used to describe the properties of the study as well as relationships of the study to other studies such as through a common Consortium. These resources can also be used to indicate participant counts associated with each of the different types of access restrictions (consent, etc.) associated with components of the study.
For this example, we have taken the Baylor Hopkins Center for Mendelian Genomics DbGaP study and built some of the relevant resources that in compliance to the Research Study Model.
ResearchConsortium
To discuss this particular set of research studies, we must first describe the role of the consortium when discussing studies of this type. The consortium, Centers for Mendelian Genomics, is used in the NCPI FHIR Model as a mechanism for aggregating member studies together. In the diagram above, you can see that, in addition to BH-CMG, there are several other centers which would each have their own set of studies. By representing the consortium in this way, we can show their relationship to one another by association to the Consortium.
The profile, ResearchConsortium, is very simple in nature. There is a single attribute that is required specific to this IG, the name. Recommended attributes include any common Acronyms and the URL for the consortium’s website.
For this example, we have created a ResearchConsortium resource, The Center for Mendelian Genomics.
{
"resourceType" : "Organization",
"id" : "cmg-research-consortium",
"meta" : {
"profile" : [
"https://nih-ncpi.github.io/ncpi-fhir-ig/StructureDefinition/ncpi-research-consortium"
]
},
"name" : "Center for Mendelian Genomics Consortium",
"alias" : [
"CMG"
],
"telecom" : [
{
"system" : "url",
"value" : "http://mendelian.org/"
}
]
}
In this compliant example, we indicate the name of the consortium using the name attribute:
"name": "The Center for Mendelian Genomics Consortium"
Since this is commonly referred to as CMG, we provide that acronym as an alias:
{
"alias": [
"CMG"
]
}
And finally, in order to provide the consortium’s website, we provide it using the telecom attribute:
{
"telecom": [
{
"system": "url",
"value": "http://mendelian.org"
}
]
}
The system, in this case, is how consumers of this data can properly interpret the value as a website URL.
NCPI Research Study, Baylor Hopkins, CMG
In this example, we have a dbGaP study, BH-CMG, which has two ways in which participants were consented. We’ve represented this main study as an NCPI Research Study and will represent each of those consent components as Research Study Subject resources.
First, let’s take a look at the study itself:
{
"resourceType" : "ResearchStudy",
"id" : "cmg-research-study-bhcmg",
"meta" : {
"profile" : ["https://nih-ncpi.github.io/ncpi-fhir-ig/StructureDefinition/ncpi-research-study"]
},
"identifier" : [{
"system" : "http://mendelian.org/fhir",
"value" : "BH-CMG"
},
{
"system" : "https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=",
"value" : "phs000711"
}],
"title" : "Baylor Hopkins Center for Mendelian Genomics (BH CMG)",
"status" : "completed",
"category" : [{
"coding" : [{
"system" : "https://nih-ncpi.github.io/ncpi-fhir-ig/CodeSystem/ncpi",
"code" : "StudyCohort",
"display" : "Study Cohort"
}]
}],
"focus" : [{
"coding" : [{
"system" : "http://purl.obolibrary.org/obo/mondo.owl",
"code" : "MONDO:0016054",
"display" : "cerebral malformation"
}]
},
{
"coding" : [{
"system" : "http://purl.obolibrary.org/obo/hp.owl",
"code" : "HP:0030853",
"display" : "Heterotaxy"
},
{
"system" : "urn:oid:2.16.840.1.113883.6.177",
"code" : "D059446",
"display" : "Heterotaxy Syndrome"
}]
},
{
"coding" : [{
"system" : "http://purl.obolibrary.org/obo/mondo.owl",
"code" : "MONDO:0016033",
"display" : "Cornelia de Lange syndrome"
}]
}],
"relatedArtifact" : [
{
"type" : "derived-from",
"label" : "DbGaP",
"url" : "phs000711"
},
{
"type" : "documentation",
"label" : "Citation",
"citation" : "Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35-61. doi: 10.1146/annurev-med-051010-162644. PMID: 22248320; PMCID: PMC3656720.",
"document" : {
"url" : "https://pubmed.ncbi.nlm.nih.gov/22248320/"
}
},
{
"type" : "documentation",
"label" : "Website",
"document" : {
"url" : "https://mendeliangenomics.org/"
}
}
],
"description" : "The Centers for Mendelian Genomics project uses next-generation sequencing and computational approaches to discover the genes and variants that underlie Mendelian conditions. By discovering genes that cause Mendelian conditions, we will expand our understanding of their biology to facilitate diagnosis and new treatments.",
"enrollment" : [{
"reference" : "Group/cmg-research-study-bhcmg-group"
}],
"sponsor" : {
"reference" : "Organization/cmg-research-consortium"
},
"extension" : [{
"url" : "https://nih-ncpi.github.io/ncpi-fhir-ig/StructureDefinition/research-study-design",
"valueCodeableConcept" : {
"coding" : [{
"system" : "https://dbgap-api.ncbi.nlm.nih.gov/fhir/x1/CodeSystem/ResearchStudy-StudyDesign",
"code" : "Mendelian",
"display" : "Mendelian"
},
{
"system" : "https://nih-ncpi.github.io/ncpi-fhir-ig/CodeSystem/ncpi-study-types",
"code" : "Mixed",
"display" : "Mixed"
}]
}
}]
}
The first thing I’ll point out is the use of identifier:
{
"identifier" : [
{
"system" : "http://mendelian.org/fhir",
"value" : "BH-CMG"
},
{
"system" : "https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=",
"value" : "phs000711"
}
]
}
We are providing two identifiers. One to distinguish it using the more familiar identifier, BH-CMG. Next, we are proving the dbGaP Accession ID. Both of these identifiers will be reasonable ways to refer to the study, depending on the user’s relationship to the study itself.
One of the required attributes is the title. "title" : "Baylor Hopkins Center for Mendelian Genomics (BH CMG)" We have added this requirement in order to ensure that the resource will be reasonably informative to those who are using data compliant with the NCPI FHIR IG.
We defined the consortium to which this study belongs above. To associate this study to that consortium, we point the sponsor attribute to a reference to that consortium resource:
{
"sponsor": {
"reference" : "Organization/cmg-research-consortium"
}
}
This provides a way to search for research studies who are sponsored by the consortium of interest. Perhaps even more importantly, this reference is actually a valid path to the entity being referred to. In this case, it’s a local reference to the Organization, CMG Research Consortium and is a valid Path to that specific resource.
Another key component is participant data. We’ll define the actual study group below, but it is an important part of the Study and is, therefore, required. We’ve added restrictions to require enrollment to have a single StudyGroup. We’ve done this to enforce proper implementation of the model itself. If there are different combinations of participant enrollment, those should be captured as substudies whose “partOf” property points to this study. For this particular study’s enrollment, we expect that particular StudyGroup to consist of the entire population associated with the study at hand.
{
"enrollment" : [
{
"reference" : "Group/cmg-research-study-bhcmg-group"
}
]
}
As stated above, this reference should be a valid path to the relevant resource.
While it isn’t required, it is strongly recommended that each study resource have a complete and meaningful description that can be provided to researchers who may be interested in accessing the data from this study. For that, we use the description attribute: "description" : "The Centers for Mendelian Genomics project uses next-generation sequencing and computational approaches to discover the genes and variants that underlie Mendelian conditions. By discovering genes that cause Mendelian conditions, we will expand our understanding of their biology to facilitate diagnosis and new treatments.",
In addition to details about the study itself and it’s enrollment details, researches will want to be able to find information about the data that goes beyond the study itself including Citations for talks and papers that used the data, the study’s website, if one exists and other types of document URLs that may be of interest to researches interested in finding out more about this study.
For this study, we’ve added a few of these items using the relatedArtifact attribute:
{
"relatedArtifact" : [
{
"type" : "derived-from",
"label" : "DbGaP",
"display": "phs000711",
"url" : "https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000711"
},
{
"type" : "documentation",
"label" : "Citation",
"citation" : "Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35-61. doi: 10.1146/annurev-med-051010-162644. PMID: 22248320; PMCID: PMC3656720.",
"document" : {
"url" : "https://pubmed.ncbi.nlm.nih.gov/22248320/"
}
},
{
"type" : "documentation",
"label" : "Website",
"document" : {
"url" : "https://mendeliangenomics.org/"
}
}
]
}
With these 3 artifacts, we provide the dbGaP study link and related ID, one of the citations that reference the study and the study’s official website.
One of the important features for making these studies findable is annotating them with proper disease codes relating the study to those diseases and phenotypes that are under study. In this case, we are only representing 3, but there is no reason to stop with any particular number. For the purpose of being concise, we’ll look at one of those three diseases:
{
"focus" : [
{
"coding" : [{
"system" : "http://purl.obolibrary.org/obo/hp.owl",
"code" : "HP:0030853",
"display" : "Heterotaxy"
},
{
"system" : "urn:oid:2.16.840.1.113883.6.177",
"code" : "D059446",
"display" : "Heterotaxy Syndrome"
}]
}
]
}
For this one, we’ve included codings from two different ontologies: HPO and MeSH relating to Heterotaxy granting end users greater flexibility in which terms they choose for their searches.
Before getting into the individual sub-studies which represent the different types of consent participants agreed to, we should discuss a few related resources.
Access Control - ResearchStudyConsent
For the purposes of this implementation guide, when we talk about access control, we are not discussing restricting access to data within FHIR, but about the limitations associated with the research data itself as is defined by the study’s principal investigators. There are expected to be appropriate controls in place to restrict access to the data according to those restrictions, but that is not up to the IG to determine how that happens.
What we can discuss is what those restrictions are so that FHIR resource consumers can identify whether the data being described can be used in their research. Once a researcher identifies the data that meets their needs, it is beyond the scope of this IG to determine how they go about getting access to that data.
While there are likely many ways to describe access control, one of the more common mechanisms used in research is that of consent and a common approach is to aggregate participants together according to the consent they agreed upon. In FHIR, we represent this consent using the profile, Research Study Consent. Our BM-CMG example contains two different consent resources: HMB-NPU and HMB-IRB-NPU.
Because they are very similar, we’ll look at one of them in detail. It is important to note that a study may consent participants differently as the study progresses and that each of those different consent configurations should be represented in FHIR.
{
"resourceType" : "Consent",
"id" : "hmb-irb-npu-consent",
"meta" : {
"profile" : [
"https://nih-ncpi.github.io/ncpi-fhir-ig/StructureDefinition/research-study-consent"
]
},
"status" : "draft",
"scope" : {
"coding" : [
{
"code" : "research"
}
]
},
"category" : [
{
"coding" : [
{
"system" : "http://terminology.hl7.org/CodeSystem/consentcategorycodes",
"code" : "research",
"display" : "Research Information Access"
}
]
}
],
"provision" : {
"type" : "permit",
"purpose" : [
{
"system" : "https://nih-ncpi.github.io/ncpi-fhir-ig/CodeSystem/nih-consent",
"code" : "HMB",
"display" : "Health/Medical/Biomedical"
},
{
"system" : "https://nih-ncpi.github.io/ncpi-fhir-ig/CodeSystem/nih-consent",
"code" : "IRB",
"display" : "IRB approval required"
},
{
"system" : "https://nih-ncpi.github.io/ncpi-fhir-ig/CodeSystem/nih-consent",
"code" : "NPU",
"display" : "Not-for-profit use only"
}
]
}
}
The most important thing to note with this resource is that of the provision:
{
"provision" : {
"type" : "permit",
"purpose" : [
{
"system" : "https://nih-ncpi.github.io/ncpi-fhir-ig/CodeSystem/nih-consent",
"code" : "HMB",
"display" : "Health/Medical/Biomedical"
},
{
"system" : "https://nih-ncpi.github.io/ncpi-fhir-ig/CodeSystem/nih-consent",
"code" : "IRB",
"display" : "IRB approval required"
},
{
"system" : "https://nih-ncpi.github.io/ncpi-fhir-ig/CodeSystem/nih-consent",
"code" : "NPU",
"display" : "Not-for-profit use only"
}
]
}
}
Each of those member codes represent a different facet of the consent which, together, represents the complete set of restrictions put into place for researchers intending on using the data for their own research. In this case, researchers can only use this data for health, medical or biomedical research at a non-profit organization after approval by the IRB panel.
Participant Groups - StudyGroup
In FHIR, we use the StudyGroup profile to establish the population under study. At the top-most level, the StudyGroup would point to the entire population of the research study. For sub-studies such as consent groups, the StudyGroup reflects only the relevant members of that grouping.
The groups provide two main pieces of information:
- Group Population
- Group Size
The Population itself is only relevant if there is a desire to link the group to the actual participants, such as when the server is hosting Study details as well as patient level data. However, if the purpose of the FHIR server is only to provide information about the studies for the purposes of findability, the population itself is no required to be enumerated. Patients will be enumerated using the member attribute in when linking to the patient data is relevant.
Group size, on the other hand, is a must. This detail makes it possible to summarize the contents of a study without having access to the row level data. For the purposes of these examples, we’ll only provide the group size.
StudyGroup - Examples
For this particular example, we have 3 different StudyGroups that we need to define. One for the entire study population and 1 for each of the two Consents that we have chosen to represent. These are very similar, so we’ll only cover one of the three, but users should feel free to look at each of them: BM-CMG Study Partipants, BM-CMG Consented by HMB-NPU and BH-CMG Consented by HMB-IRB-NPU.
Let’s take a look at the HMB-IRB StudyGroup:
{
"resourceType" : "Group",
"id" : "cmg-research-study-bhcmg-consent-group-hmb-irb-npu",
"meta" : {
"profile" : [
"https://nih-ncpi.github.io/ncpi-fhir-ig/StructureDefinition/study-group"
]
},
"type" : "person",
"actual" : true,
"name" : "BM-CMG Study HMB-IRB-NPU Participants",
"quantity" : 780
}
As you can see, there isn’t too much there because this example isn’t intended to refer to patient level data. As such, we opted to leave the member attribute out of the resource.
The first and most important thing to note is the quantity: "quantity": 780. As mentioned above, this is vital for summary purposes since the actual membership isn’t required. We also require the use of "actual": true which indicates that the group is describing a specific set of participants as opposed to being a descriptive set of characteristics. The profile also requires that the group’s type be that of person since our current focus is on Human Research. We indicate this using the type as follows: "type": "person".
While it isn’t required, we strongly recommend using meaningful names to help clarify to the consumer what the group is for when seen outside the context of other, related resources. In this case, we see the following: "name": "BM-CMG Study HMB-IRB-NPU Participants"
Research Study Subject
While there may be many reasons to tie a subset of the study’s entire population to a sub-study, such as called whole genomes on a sub-population due to budget, the focus of this profile is to distinguish the members of a common set of data restrictions. Often, this is referred to as the Consent group since the group of participants are consented with the same set of data restrictions. To represent these groups in FHIR, we are modelling the main component of this top level R4 resource, the ResearchStudySubject, after the R5 version of FHIR’s ResearchSubject resource. This allows us to tie a single Consent resource to the group and to associate it with the parent ResearchStudy using the partOf attribute.
For the purposes of this example, we’ve represented two of the consent groups listed in dbGaP for BM-CMG, HMB-NPU and HMB-IRB-NPU. These two resources are very similar, so we’ll only discuss one of them in detail.
Research Study Subject, BH-CMG HMB-IRB-NPU
As stated above, the chief goal of the ResearchStudySubject is to associate a portion of Participants whose data is restricted in identical ways. In this case, by the restrictions, Health/Medical/Biomedical Research, “IRB approval required” and Not-for-profit use only. These restrictions specify the limits any researcher is bound to when using the data consented in this way.
{
"resourceType" : "ResearchStudy",
"id" : "cmg-research-study-subject-cmg-hmb-irb-npu",
"meta" : {
"profile" : [
"https://nih-ncpi.github.io/ncpi-fhir-ig/StructureDefinition/research-study-subject"
]
},
"extension" : [
{
"url" : "https://nih-ncpi.github.io/ncpi-fhir-ig/StructureDefinition/research-study-subject-consent",
"valueReference" : {
"reference" : "Consent/hmb-irb-npu-consent"
}
}
],
"identifier" : [
{
"system" : "http://mendelian.org/fhir",
"value" : "BH-CMG-HMB-IRB-NPU"
}
],
"title" : "BH-CMG HMB-IRB-NPU Consent Study Subject",
"partOf" : [
{
"reference" : "ResearchStudy/cmg-research-study-bhcmg"
}
],
"status" : "completed",
"enrollment" : [
{
"reference" : "Group/cmg-research-study-bhcmg-consent-group-hmb-irb-npu"
}
]
}
The first thing to note is the reference to the parent study by way of the partOf attribute:
{
"partOf" : [
{
"reference" : "ResearchStudy/cmg-research-study-bhcmg"
}
]
}
This ties the other StudyGroup associated with this portion of the study back to the parent study.
Next notice the reference to the appropriate StudyGroup in the enrollment attribute:
{
"enrollment" : [
{
"reference" : "Group/cmg-research-study-bhcmg-consent-group-hmb-irb-npu"
}
]
}
This links the ResearchStudySubject to the StudyGroup which describes the participants consented under the HMB, IRB and NPU consent.
Finally, we link the link to the actual consent resource using the extension, research-study-subject-consent.
{
"extension" : [
{
"url" : "https://nih-ncpi.github.io/ncpi-fhir-ig/StructureDefinition/research-study-subject-consent",
"valueReference" : {
"reference" : "Consent/hmb-irb-npu-consent"
}
}
]
}
This is a feature of the R5 version of ResearchStudy that allows us to match all members of the one StudyGroup to a single consent.